-
×
 BGWR3001 Baggywel - Eau Peptonee Tamponnee - 2 X 5l Alliance Bio Exterpise
1 × $32.50
BGWR3001 Baggywel - Eau Peptonee Tamponnee - 2 X 5l Alliance Bio Exterpise
1 × $32.50 -
×
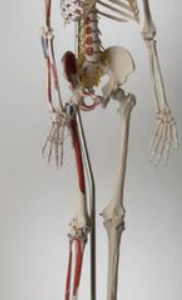 S58L Premier Academic Kinesiology Skeleton, Painted And Labeled, Sacral Mount On Mobile Stand- Denoyer Geppert
2 × $2,747.00
S58L Premier Academic Kinesiology Skeleton, Painted And Labeled, Sacral Mount On Mobile Stand- Denoyer Geppert
2 × $2,747.00 -
×
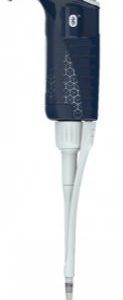 F81043-PIPETMAN M P200M BT Connected-Gilson
1 × $907.00
F81043-PIPETMAN M P200M BT Connected-Gilson
1 × $907.00 -
×
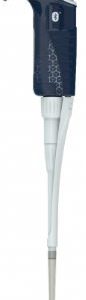 F81045-PIPETMAN M P1200M BT Connected-Gilson
1 × $907.00
F81045-PIPETMAN M P1200M BT Connected-Gilson
1 × $907.00 -
×
 FA10005M-PIPETMAN L P200L, 20-200 µL, Metal Ejector-Gilson
1 × $499.00
FA10005M-PIPETMAN L P200L, 20-200 µL, Metal Ejector-Gilson
1 × $499.00 -
×
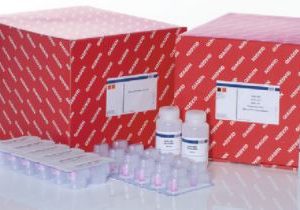 74116-RNeasy Mini QIAcube Kit (240)- Qiagen
1 × $2,627.00
74116-RNeasy Mini QIAcube Kit (240)- Qiagen
1 × $2,627.00 -
×
 BSH100-02 Block, 40 x 0.2ml (or 5 x PCR strips) Benchmark Scientific
2 × $87.00
BSH100-02 Block, 40 x 0.2ml (or 5 x PCR strips) Benchmark Scientific
2 × $87.00 -
×
 F81046-PIPETMAN M P5000M BT Connected-Gilson
1 × $907.00
F81046-PIPETMAN M P5000M BT Connected-Gilson
1 × $907.00 -
×
 3970302- 3' Purifier Filtered PCR Enclosure with UV Light and Protection Panel- Labconco
1 × $9,062.00
3970302- 3' Purifier Filtered PCR Enclosure with UV Light and Protection Panel- Labconco
1 × $9,062.00 -
×
 BSWCMB Quick-Flip Block Benchmark Scientific
1 × $154.00
BSWCMB Quick-Flip Block Benchmark Scientific
1 × $154.00 -
×
 BSH100-1213 Block, 15 x 12/13mm diameter tubes & vacutainers Benchmark Scientific
3 × $99.00
BSH100-1213 Block, 15 x 12/13mm diameter tubes & vacutainers Benchmark Scientific
3 × $99.00 -
×
 F144075-PIPETMAN G Multichannel P12x300G, 30-300µL-Gilson
2 × $1,044.00
F144075-PIPETMAN G Multichannel P12x300G, 30-300µL-Gilson
2 × $1,044.00 -
×
 M201-Pregnancy Experience Suit-Sakamoto
1 × $1,004.00
M201-Pregnancy Experience Suit-Sakamoto
1 × $1,004.00 -
×
 4316 Polypropylene 0.5ml Graduated Flat Top Microcentrifuge Tube, Red Bio Plas
1 × $12.00
4316 Polypropylene 0.5ml Graduated Flat Top Microcentrifuge Tube, Red Bio Plas
1 × $12.00 -
×
 F81040-PIPETMAN M P10M BT Connected-Gilson
2 × $907.00
F81040-PIPETMAN M P10M BT Connected-Gilson
2 × $907.00 -
×
 BSWNMR Block, 30 x NMR tubes Benchmark Scientific
1 × $227.00
BSWNMR Block, 30 x NMR tubes Benchmark Scientific
1 × $227.00 -
×
 BCLO3016 Paper Roll For Bluetooth Printer Bas Alliance Bio Expertise
2 × $42.50
BCLO3016 Paper Roll For Bluetooth Printer Bas Alliance Bio Expertise
2 × $42.50
Subtotal: $26,290.50





