AL101 – MIM-C10 Phage Display Peptide Library Kit -Antibody Design
$973.50
Technical Description
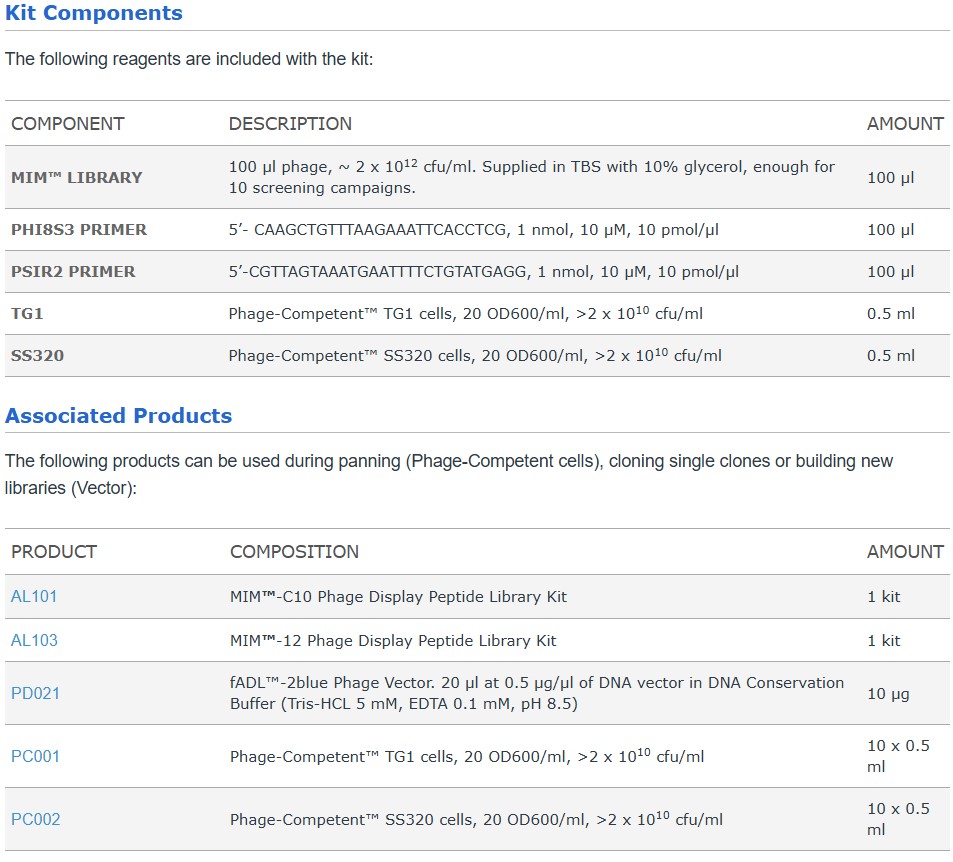
The MIM™-C10 Phage Display Peptide Library consists of ~109 random cyclic 10-mer peptides displayed as an N-terminal fusion protein with the minor coat protein pIII on the head of the fd filamentous bacteriophage. The sequence of the insert is NH2-AEGCX10C GGGSGPGGLRGGS-pIII after cleavage of the leader peptide. The library was built on the phage vector fADL-2blue using trimer codon technology where unwanted stop codons and cysteines are absent. fADL™-2blue is a multivalent type 3 phage display vector (Smith 1997) derived from the phage vector fd-tet (Zacher 1980). fADL-2blue is well tolerated by the host and gives large colonies and small plaques, simplifying the screening of libraries and the handling of single clones. Helper phage are not required for amplification. The linker GGGSGPGGLRGGS is sensitive to trypsin, enabling trysin elution, which has been show to improve motif recovery (Thomas 2010).
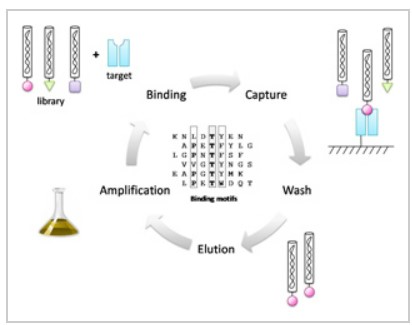
Figure 1. Principle of library selection through iterative rounds of biopanning.
The MIM™-C10 Phage Display Peptide Library Kit holds enough material for 10 panning experiments (Fig. 1). Each aliquot of 10 µl contains 2 x 1010 cfu, equivalent 20 transducing units per peptide on average. Panning of the library leads to the rapid identification of peptide binders (Fig. 2).

Figure 2. M2 Antibody Epitope Mapping. Panning of the MIM-C10 library by the FLAG® tag M2 antibody leads to >90% sequences with motifs matching the tag sequence on the third round.
Applications
-
Epitope mapping.
-
De novo isolation of peptide binders.
-
Identification of peptide binding motifs.
Quality Control & Certification of Analysis
Quality Control Assays
Specifications for each lot are reported in the certificate of analysis.
Certification
Product meet all specifications.